Multi-scale Community Finder
Network has become a popular means to model complex relationships in biological systems, examples include genome-wide co-expression studies, gene regulatory networks, and protein-protein interaction networks etc. Often, these networks require clustering analysis, in which groups of densely connected nodes are identified. In fact, modular structure is deemed important characteristics in biological networks. Unlike traditional clustering methods, communities (i.e., clusters) in network representation are subject to ‘resolution limit’, which means some smaller communities cannot be detected by simply optimizing the modularity measure. This overlook may cause inaccurate or misleading functional annotations of groups of nodes based on modular structures of networks. Considering this, statistical methods were developed to deal with the multi-scale community profiles in complex networks. In particular, Mucha et al. proposed a systemic approach to unfold multi-scale multiplex community structures. In multiplex networks that involve multiple time or context dependent networks slices, the same controlling parameter (referred to as the scale of the community profile) in single-slice networks is generalized to multi-slice networks. Such advance in community detection is useful in studying many biological problems in system biological paradigms, including the study of time-coursed data and integrative network analysis of high-throughput data.
Existing tools in finding network communities (a.k.a. graph clusters) such as ‘jClust’ and ‘GLay’ only implement graph clustering methods without consideration of multiple scales. ‘igraph’ include a primary version multi-scale community detection method for only undirected networks. Here we developed a fast tool, Multi-scale Community Finder (MCF), based on modularity improvement heuristic in finding multi-scale community structures in all major types of networks, including (un)directed, signed, bipartite, multi-slice networks. We implemented two different methods for controlling scales of networks from recent studies.
Director
Developers
Shang Gao, Alan Chen, Ali Rahmani, Tamer N. Jarada
Related papers
Related papers are listed under the website of Dr. Reda Alhajj
Copyright
The copyright and proper citation is required if the tool is to be used.

- Software:
- Testing samples:
Multi-scale Community Finder Tutorial & File(s) Types - click on one of these choices
- Download CommunityFinder.zip
- Start CommunityFinderUI.exe
- Select tab representing the network’s time series:
- Single-Slice: Snapshot of the network at one time period
- Multi-Slice: Snapshots of the network across multiple time periods
-
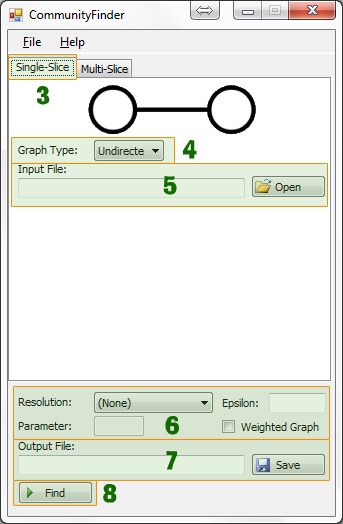
Single-Slice
- Select graph type
- Select input file (network links file)
- Set optional parameters:
- Modularity (bi-partite graphs only)
- Resolution type.
- R value
- Gamma
- Epsilon
- Select where to save output file (communities file)
- Click Find
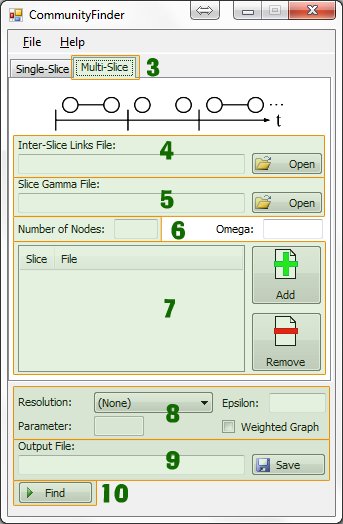
Multi-Slice
- Select inter-slice links file
- Select slice gamma file
- Set number of nodes in the network (number of unique nodes)
- Add in all slice files chronologically (network links files at each time slice)
- Set optional parameters:
- Omega
- Resolution type
- R value
- Gamma
- Epsilon
- Select where to save output file (communities file)
- Click Find
CommunityFinder will accept two different types of input files for network links:
- Text files (*.txt)
- Comma-separated values files (*.csv)
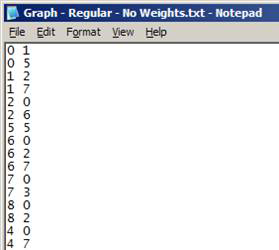
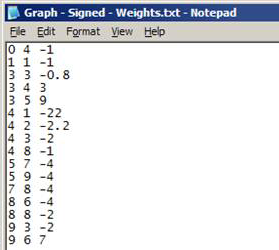
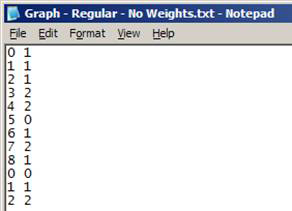
Text file (*.txt)
- No headers
- Each row represents a link in the network:
- Format: origin_node destination_node [link_weight]
- link_weight is optional
- Nodes must be numbered from 0 … (n – 1)
Note: This format is preferred for large networks
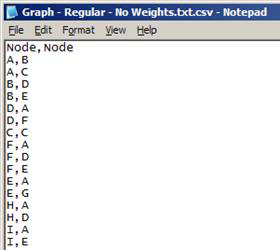
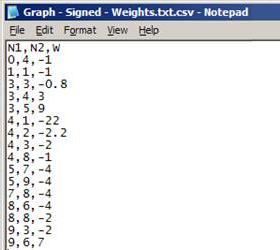
Comma-separated values file (*.csv)
- First row contains the headers (any values for headers are acceptable)
- Each row represents a link in the network:
- Format: origin_node,destination_node, [link_weight]
- link_weight is optional
- Nodes can have any values for names
CommunityFinder will accept two different types of inter-slice links files:
- Text files (*.txt)
- Comma-separated values files (*.csv)
- File type needs to match Slice Files’ type chosen
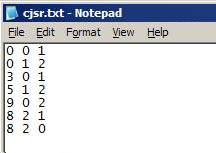
Text file (*.txt)
- No headers
- Each row represents a link across slices in the network:
- Format: node origin_slice_number destination_slice_number
- Nodes must be numbered from 0 … (n – 1)
- Slice numbers are numbered from 0
- Numbers refer to the order in which Slice Files are added
Note: This format is preferred for large networks
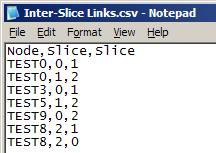
Comma-separated values file (*.csv)
- First row contains the headers (any values for headers are acceptable)
- Each row represents a link across slices in the network:
- Format: node,origin_slice_number,destination_slice_number
- Nodes can have any values for names
- Slice numbers are numbered from 0
- Numbers refer to the order in which Slice Files are added
CommunityFinder will accept two different types of slice gamma files:
- Text files (*.txt)
- Comma-separated values files (*.csv)
- File type needs to match Slice Files’ type chosen
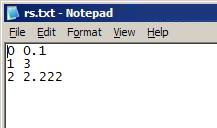
Text file (*.txt)
- No headers
- Each row indicates a gamma to apply for a slice in the network:
- Format: slice_number gamma
- Slice numbers are numbered from 0
- Numbers refer to the order in which Slice Files are added
Note: This format is preferred for large networks
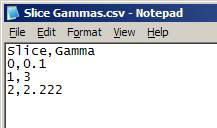
Comma-separated values file (*.csv)
- First row contains the headers (any values for headers are acceptable)
- Each row indicates a gamma to apply for a slice in the network:
- Format: slice_number,gamma
- Slice numbers are numbered from 0
- Numbers refer to the order in which Slice Files are added
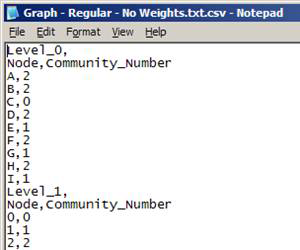
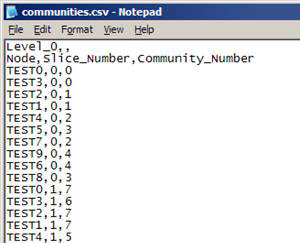
CommunityFinder can generate two different types of output files for communities:
- Text files (*.txt)
- Comma-separated values files (*.csv)
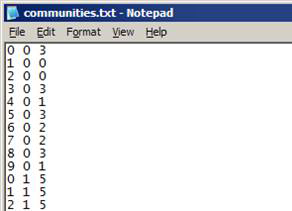
Text file (*.txt)
- No headers
- Each row represents a node and it’s assigned community:
- Format: node [slide_number] community_number
- Slide_number only appears for the first level of multi-slice networks
- Communities are numbered from 0 … (n – 1)
- Start of next level of communities is identified by node 0
Note: This format is preferred for large networks
Comma-separated values file (*.csv)
- Two sets of headers at each level:
- Indicates level number starting at 0
- Indicates column values
- Each row represents a node and it’s assigned community:
- Format: origin_node,[slide_number],destination_node
- Slide_number only appears for the first level of multi-slice networks
- Communities are numbered from 0 … (n – 1)
- Start of next level of communities is identified by node 0